Inference in Bayesian Networks¶
Inference¶
Inference Problem:
- Given: a Bayesian network
- Given an assignment of values to some of the variables in the network: E_i=e_i(i=1,\dots,l)
- "Instantiation of the nodes \bold E"
- "Evidence \bold E=e entered"
- "Findings entered"
- ...
- Want: for variables A\notin \bold E the posterior margnial P(A\mid \bold E=e)
According to the definition of conditional probability, it is sufficient to compute for each A\in D_A the value
Together with
This gives the posterior distribution
Inference as Summation¶
Let A be the variable of interest, \bold E the evidence variables, and \bold Y=Y_1,\dots,Y_l the remaining variables in the network not belonging to A\cup \bold E. Then
$$ P(A=a,\bold E=e)=\sum_{y_1\in D_{Y_1}}\cdots\sum_{y_l\in D_{Y_l}}P(A=a,\bold E=e,Y_1=y_1,\dots,Y_l=y_l) $$ Note:
- For each \bold y the probability P(A=a,\bold E = \bold e, \bold Y = \bold y) can be computed from the network In time linear in the number of random variables
- The number of configurations over \bold Y is exponential in l
Inference Problems¶
First Problem¶

Find P(B\mid a,f,g,h)={P(B,a,f,g,h)\over P(a,f,g,h)}
We can if we have access to P(A,B,C,D,E,F,G,H)
Inserting evidence we get:

and

Second Problem¶

See naive solution in Lecture 14.10 Slides 8-11
Naive Solution Summary¶

Variable Elimination¶
Problem
The joint probability distribution will contain exponentially many entries
Idea
We can use
- the form of the joint distribution P, and
- the law of distributivity
to make the computation of the sum more efficient
Thus, we can adapt our elimination procedure so that:
- we marginalize out variables sequentially
- when marginalizing out a particular variable X, we only need to consider the factors involving X
Example¶



Factors¶
Calculus of factors
-
The procedure operates on factors: functions of subsets of variables
-
Required operations on factors:
- multiplication
- marginalization (summing out selected variables)
- restriction (setting selected variables to specific values)
Complexity

See example in Lecture 14.10 Slides 16-17
Singly Connected Networks¶
A singly connected network is a network in which any two nodes are connected by at most one path of undirected edges.
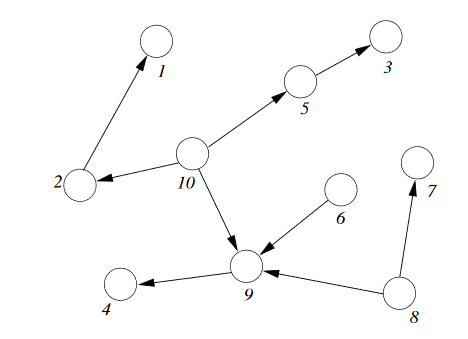
For singly connected network: any elimination order that "peels" variables from outside will only create factors of one variable.
The complexity of inference is therefore linear in the total size of the network ( = combined size of all conditional probability tables)
Approximate Inference¶
Sample Generator¶
Observation: can use Bayesian network as random generator that produces states \bold X = \bold x according to distribution P defined by the network
Example¶

Approximate Inference from Samples¶
To compute an approximation of P(\bold E = \bold e) (\bold E is a subset of the variables in the Bayesian network):
- generate a (large) number of random states
- count the frequency of states in which \bold E = \bold e

Hoeffding Bound¶

Required Sample Size¶

Example¶


Rejection Sampling¶
The simplest approach: Rejection Sampling

Problem
Samples with \bold E \neq \bold e are useless!
Ideally: would draw samples directly from the conditional distribution P(\bold A \mid \bold E = \bold e)

Likelihood Weighting¶
We would like to sample from

So instead weigh each generated sample with a weight corresponding to Part 2

